Welcome to the Atkinson Lab at Baylor College of Medicine!
The Atkinson Lab develops statistical genomics methods that account for the full range of human population structure and genetic variation. We use globally representative genomic datasets and cutting-edge computational approaches to study complex neuropsychiatric traits, with a particular focus on populations with admixed ancestry. Our tools are broadly applicable across phenotypes and cohorts, supporting more accurate and generalizable inferences in human genetics. We also investigate the demographic and evolutionary history of human populations to inform downstream analytic methods.
What We Work On
Our lab investigates a range of topics in statistical and human genetics to improve the accuracy and applicability of genomic studies across globally representative populations. We have three primary research tracks: (1) applied and analytic work in statistical genetics, (2) investigations into human population structure and evolutionary history, and (3) development of computational methods and tools for large-scale genomic analysis.
Statistical Genetics
Population Genetics
Psychiatric Genetics
Collaborative Research
Research Areas
Method Development for Ancestry-Aware Genomic Analyses
Nearly 80% of genome-wide association studies (GWAS) to date have focused on individuals of European ancestry. This has created methodological gaps in the analysis of admixed individuals—those with genomic contributions from multiple ancestral backgrounds—whose genomes present additional modeling challenges. Our lab develops statistical tools specifically designed to improve the analysis of complex traits in admixed populations, with the goal of enabling broader applicability of genomic findings, particularly in the context of mental health genetics.
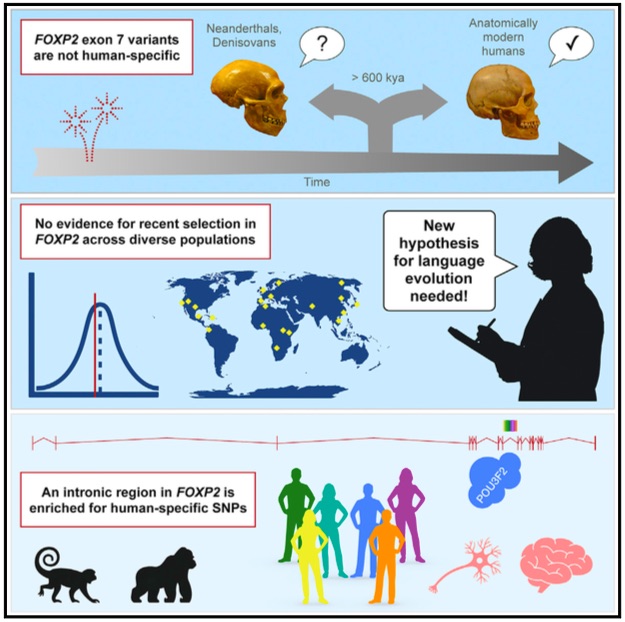
Characterizing Human Evolution and Population Structure Using Global Genomic Data
Understanding the evolutionary and demographic history of human populations is critical for interpreting patterns of genetic variation and for developing appropriate statistical and medical genomic tools. Our lab investigates these foundational aspects using ancestrally informative evolutionary statistics and globally representative DNA datasets. By characterizing the forces that shape genetic variation in key genes—including those relevant to brain function—we aim to inform method development for downstream genomic analyses across a range of populations and phenotypes.
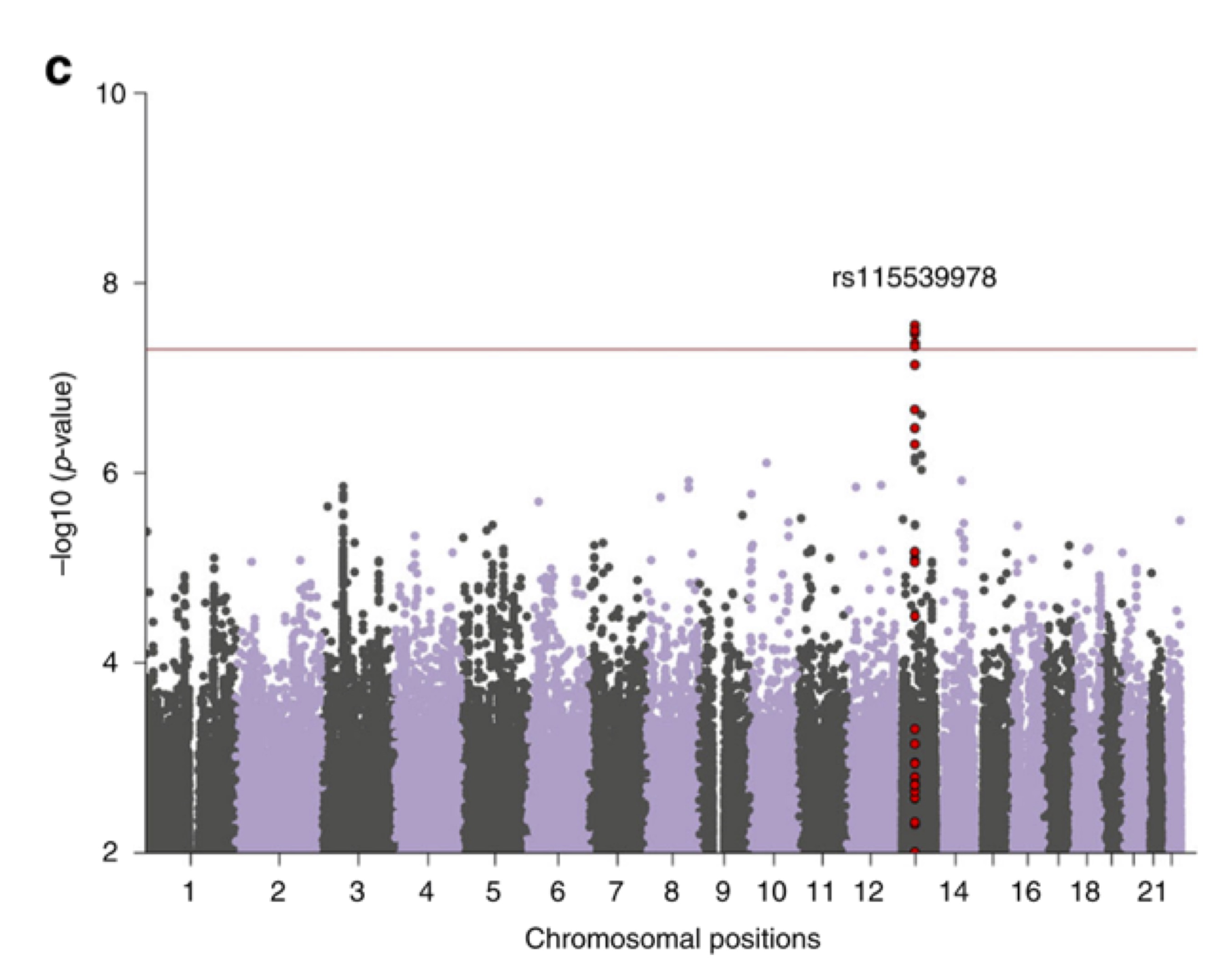
Understanding Genetic Contributors to Psychiatric Traits Across Global Populations
Psychiatric disorders are a leading global contributor to disability and represent a major focus of human genetic research. Our lab collaborates with international consortia to develop and apply statistical methods that account for population structure in the analysis of psychiatric phenotypes. These efforts aim to improve the resolution and generalizability of genetic findings related to mental health across globally representative cohorts.

Building Representative Genomic Resources for Complex Trait Research
Advancing statistical genetics requires large-scale genomic datasets that reflect global population structure and phenotypic variation. Our lab plays a leadership role in several major international collaborations—including the Psychiatric Genomics Consortium (PTSD working group), Latin American Trans-Ancestry Initiative for OCD, and the Latin American Genomics Consortium (LAGC)—which collectively support the development of more representative resources for gene discovery. These efforts also provide trainees with valuable opportunities to work with richly phenotyped, globally relevant cohorts.
Team

Astrid Manuel
Postdoctoral Fellow
As a postdoctoral associate in Dr. Elizabeth Atkinson’s lab, Astrid’s current research goals involve the development of statical genomics methods for studying complex traits in heterogeneous populations in biomedical research. She aims to apply these methods to advance pharmacogenomics and personalized medicine applications. Astrid earned her PhD from the University of Texas Health Science Center (UTHealth) in Houston, TX. The title of her PhD thesis was “Translational Bioinformatics Approaches for Decoding Mechanisms and Developing Drug Repositioning Strategies in Multiple Sclerosis”. During her PhD studies, she was a Predoctoral Fellow of the National Library of Medicine (NLM) Training Program in Biomedical Informatics and Data Science (T15LM007093). In the Atkinson Lab, Astrid seeks to extend her bioinformatics expertise to the domain of statistical population-level genomics and pharmacogenomics. During her free time, she enjoys drawing, visiting art museums, and indoor rock climbing with friends.

Erik Stricker
Postdoctoral Fellow
Erik is a postdoctoral research fellow, retrovirologist and cancer researcher currently at Baylor College of Medicine. Besides his interest in human endogenous retroviruses (HERVs) - which comprise 8% of the human genome - and their relationship to cancer, he is a passionate teacher of science and science-unrelated topics. He has been fortunate to have received the chance to acquire training at leading universities in Germany and frequently share his expertise when teaching graduate and undergraduate students. He is the recipient of multiple teaching awards and enjoys pushing the boundaries of current knowledge through research and student projects.

Estela Maria Bruxel
Former Postdoctoral Fellow
Estela Maria Bruxel is a postdoctoral research fellow in Dr. Elizabeth Atkinson’s lab. She received her master’s and Ph.D. from the Federal University of Rio Grande do Sul (Brazil), studying genetic susceptibility and pharmacogenetics of Attention Deficit/Hyperactivity Disorder. At the University of Campinas, also in Brazil, she has developed a project to identify susceptibility loci for mesial temporal lobe epilepsy (MTLE) through integrating analyses of eQTL and GWAS data and prediction algorithms to predict medication response. Additionally, she formerly received a fellowship to work in Atkinson lab to develop bioinformatic ancestry-informed techniques to study Brazilian admixed samples and improve GWAS analysis. She is a member of the Latin America Genetic Consortium and is the liaison for the Psychiatry Genetic Consortium Outreach Committee. She hopes to improve the understanding of the genetic contributors to neuropsychiatric and neurological diseases and create strategies for precision medicine treatment for such patients. Out of the lab, she loves riding mountain bike and exploring parks.

Grace Tietz
Graduate Student
Grace Tietz is a Genetics & Genomics PhD student in the lab whose primary project aims to improve genetic testing for cardiovascular disease across populations. She approaches this challenge through the lens of population-genetics and integrates information on rare variants and polygenic burden to optimize performance across ancestries. Complimenting this work, she is evaluating patient perspectives to genetic testing and is developing polygenic approaches to prioritize genetic sequencing in individuals of the Undiagnosed Diseases Network. Beyond her research, Grace is interested in genetics’ role in society, and ways that policies and healthcare practices reflect our collective perspectives on it.

Hatoon Al Ali
Graduate Student
Hatoon is a PhD student in the Genetics and Genomics program at Baylor College of Medicine. She is working on developing methods for detecting epistasis and genetic interaction in rare and common diseases using population genetics approaches. She is also interested in the incorporation of Arab/Middle Eastern populations in genomic studies. Before joining the lab, Hatoon graduated from King Abdullah University of Science and Technology with a Masters in Bioinformatics and a Bachelors in Clinical laboratory sciences from King Saud University. Outside the lab she enjoys hiking, gardening, and building with mud!

Yi-Sian "Helen" Lin
Graduate Student
Helen is a PhD student in the Atkinson Lab and is in the Genetics and Genomics program at Baylor College of Medicine. She is originally from Taiwan. She completed her MS degree in Neuroscience at National Taiwan University and was a research assistant in Bioinformatics at National Yang Ming Chao Tung University. Her research interests are optimizing genetic risk prediction for neuropsychiatric conditions across populations, pharmacogenomics, and gene x environment interactions in disease prediction. Outside the lab, she enjoys playing groovy music, watching vlogs featuring cute kids or dogs, and immersing herself in nature.

Nirav Shah
Graduate Student
Nirav is a Ph.D. candidate in the Genetics and Genomics program, where he investigates the genetic architecture of complex traits and the ancestral history of admixed individuals with biobank data, as well as the genetics of PTSD as member of the Psychiatric Genomics Consortium. Before starting graduate school, he worked as a computational biologist in a Duke spinoff biotech startup in North Carolina. In his free time, he loves to explore the outdoors, and enjoys hiking, swimming and reading.

Pragati Kore
Graduate Student
Pragati is a PhD student in the Genetics & Genomics program at Baylor College of Medicine. She is broadly interested in developing and applying approaches to study rare variants with the incorporation of admixed populations. She grew up in Austin, Texas, and graduated from UT Austin with a BS in Biochemistry. Following her bachelor’s degree, she worked at the Center for Advanced Genomics Technology at the Icahn School of Medicine at Mount Sinai as an Associate Researcher. Outside of the lab, she enjoys attending concerts, traveling, and trying new recipes in the kitchen!

Jessica Honorato Mauer
Bioinformatics Analyst
Jessica was a former visiting Master's student in the Atkinson Lab, who recently returned as a bioinformatics analyst! They graduated from Universidade Federal de São Paulo, Brazil in 2019 with a BSc. in Biomedical Science, and in 2022 with a MSc. in Human Genetics/Bioinformatics. Jessica is interested in facilitatig the study of underrepresented populations in genetics research and improving disease risk prediction for heterogeneous ancestry/admixed populations. Outside of the lab, they enjoy playing games, drawing, and playing guitar.
Former Members and Friends of the Lab

Aishi Ayyanathan
Undergraduate
Aishi is an undergraduate student from Rice University who is majoring in biology and sociology. She is currently working on running genome-wide association studies for the All of Us dataset, in order to evaluate the performance of various genetic inference algorithms across population definitions. Broadly, she is interested in leveraging large-scale datasets to increase the utility of genomics research for translation. Outside of the lab, she enjoys crocheting and playing Anagrams.

Amira Ellison
Former Post-baccalaureate Scholar
Amira was a post-baccalaureate student in the Human Genome Sequencing Center Pre-Graduate Education and Training (PGET) program at Baylor College of Medicine. She completed her undergraduate study in Biology with a concentration in genetics at Pennsylvania State University in 2022. Amira’s prior research experience includes studying the gene arrangements of Klamath and Persimilis drosophila, specifically focusing on their breakpoint regions. Currently, she is working to improve polygenic risk score performance for lipoprotein(a), breast cancer, and epilepsy across ancestries.

Shantal Taylor
Former Post-baccalaureate Scholar
Shantal is an undergraduate student majoring in Chemistry with a Biomedical focus at Prairie View A&M University, Texas. She is a member of the Texas Undergraduate Medical Academy and her university’s Honors Program, and works as a student researcher in our Cooperative Agricultural Research Center. She aspires to become a primary care physician and infectious disease researcher, and is interested in exploring the vector-transmitted diseases prevalent in understudied, tropical regions. Shantal participated in the Baylor SMART program to expand her biomedical and health-related research knowledge, through which she worked in the Atkinson lab for one summer conducing a project exploring the population genetics of varied groups in the UK biobank.

Taotao Tan
Former Bioinformatics Analyst
Taotao completed his bachelor's and master’s degree in biology, and then joined the Atkinson lab as a bioinformatics analyst in 2021. His primary focus of work revolved around the statistical development and application of GWAS methodologies for admixed populations. Starting in the fall of 2023, he is pursuing his Ph.D. in the Quantitative and Computational Biosciences program at Baylor College of Medicine. In his spare time, he enjoys playing video games and the electric guitar.
Select Publications
Given our lab's high level of productivity, publications are no longer being actively updated. For the most complete and up to date list, see Dr. Atkinson's Google scholar page.
- Ragsdale, AP, Weaver, TD, Atkinson EG,Hoal EG, Möller M, Henn BM, Gravel S. (2023) A weakly structured stem for human origins in Africa. Nature., ePub ahead of print. [Online] [In popular press]
- Taotao Tan &Atkinson EG. (2023) Strategies for the Genomic Analysis of Admixed Populations. Annual Review of Biomedical Data Science, 53:195–204. [Online]
- Atkinson EG. (2023) Estimation of cross-ancestry genetic correlations within ancestry tracts of admixed samples. Nature Genetics, 55, 527–529. [Online]
- Atkinson EG. Bianchi, S.B., Ye, G.Y. et al. (2023) Cross-ancestry genomic research: time to close the gap. Neuropsychopharmacology., 47, 1737–1738. [Online]
- Majara L, Kalungi A, Koen, N ... Atkinson EG, Martin A. (2023). Low and differential polygenic score generalizability among African populations due largely to genetic diversity. Human Genetics and Genomics Advances., 4(2) 100184. [Online]
- Atkinson JG &Atkinson EG. (2023) Machine Learning and Health Care: Potential Benefits and Issues. Journal of Ambulatory Care Management, 55:527-529. [Online]
- Atkinson EG. Bianchi, S.B., Ye, G.Y. et al. (2022) Genetic structure correlates with ethnolinguistic diversity in eastern and southern Africa. American Journal of Human Genetics., 109(9) 1667-1679. [Online] [In popular press]
- Trubetskoy, V., Pardiñas, A.F., Qi, T. et al. (2022) Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature., 604, 502–508. [Online]
- Atkinson EG. Choquet, H., Khor, C. C., & Wonkam, A. (2022). Improving equity in human genomics research. Communications Biology., 5(1). [Online]
- Atkinson EG, Maihofer AX, Kanai M, Martin AR, Karczewski KJ, Santoro ML, et al. (2021) Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nature Genetics, 53:195–204. [Online][Press release][In the news]
- Atkinson EG, Audesse AJ, Palacios J, Smith GT, Bobo DM, Webb AE, Ramachandran S, Henn BM (2018). No Evidence for Recent Selection at FOXP2 among Diverse Human Populations. Cell, 174(6): 1424-1435. PMC6128738. [Online][In the news] [In popular press]
- Camarena B, Atkinson EG, Baker M, Becerra-Palars C, Chibnik LB, Escamilla-Orozco R, Jiménez-Pavón J, Koenig Z, Márquez-Luna C, Martin AR, Morales-Cedillo IP, Olivares AM, Ortega-Ortiz H, Rodriguez-Ramírez AM, Saracco-Alvarez R, Basaldua RE, Sena BF, Koenen K.C. (2021) Neuropsychiatric Genetics of Psychosis in the Mexican Population: A Genome-Wide Association Study Protocol for Schizophrenia, Schizoaffective, and Bipolar Disorder Patients and Controls. Complex Psychiatry. https://www.karger.com/Article/Abstract/518926. [Online]
- Gopalan S, Atkinson EG, Buck LT, Weaver TD, Henn BM. (2021) Inferring archaic introgression from hominin genetic data. Evolutionary Anthropology. https://doi.org/10.1002/evan.21895. [Online]
- Martin AR, Atkinson EG, Chapman SB, Stevenson A, Stroud RE, et al. (2021) Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet., 108:656-668. [Online]
- The COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19 by worldwide meta-analysis. (2021) Nature. [Online]
- Wendt FR, Pathak GA, Overstreet C, Tylee DS, Gelernter J, Atkinson EG, Polimanti, R. (2021) Characterizing the effect of background selection on the polygenicity of brain-related traits. Genomics, 113:111–119.[Online]
- Guindo-Martínez M, Amela R, Bonàs-Guarch S, Puiggròs M, Salvoro C, Miguel-Escalada I, et al. (2021) The impact of non-additive genetic associations on age-related complex diseases. Nature Communications, 12:2436.[Online]
- Scelza BA, Atkinson EG, Prall S, McElreath R, Sheehama J, Henn BM. (2020) The ethics and logistics of field-based genetic paternity studies. Evol Hum Sci 2020;2:1–36. [Online]
- Scelza BA, Prall SP, Swinford N, Gopalan S, Atkinson EG, McElreath R, et al. (2020) High rate of extrapair paternity in a human population demonstrates diversity in human reproductive strategies.Science Advances 6:eaay6195. [Online]
- Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, et al. (2019). International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nature Communications, 10:4558. [Online]
- Sugden LA, Atkinson EG, Fischer AP, Rong S, Henn BM, Ramachandran S. Localization of adaptive variants in human genomes using averaged one-dependence estimation. (2018) Nature Communications 9:703. [Online]
- Martin, A.R., Lin, M., Granka, J.M., Myrick, J.W., Liu, X., Sockell, A., Atkinson, EG, Werely, C.J., Möller, M., Sandhu, M.S., et al. (2017). An Unexpectedly Complex Architecture for Skin Pigmentation in Africans. Cell 171, 1340–1353.e14. [Online] [In the media] [Related popular press]
Contact
Interested in joining our group?
Candidates with training and interest in statistical genetics, population genetics, and/or psychiatric genetics across ancestries please email Dr. Atkinson.

Elizabeth Atkinson
1 Baylor Plaza
Taub Research Building, Rooms 621 (office) and 619 (lab)
Department of Molecular and Human Genetics
Baylor College of Medicine
Houston, TX 77030